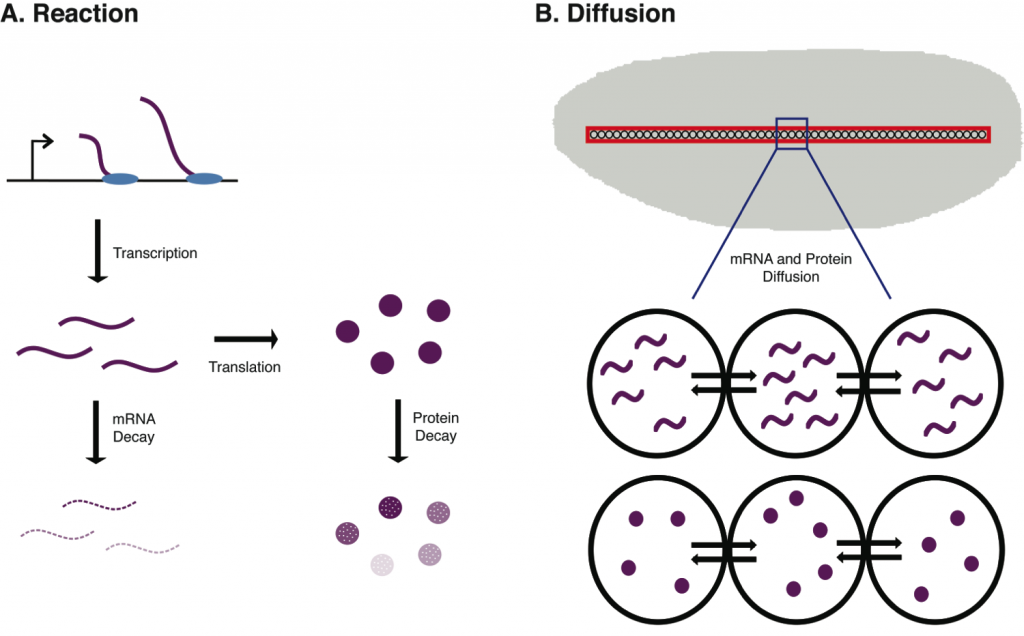
Due to the dynamic regulation of gene expression in eukaryotes, we use a set of differential equations derived from the reaction-diffusion equation to model mRNA and protein concentrations in developing Drosophila embryos. These models incorporate many important features of the biological system, including rates of transcription and translation, as well as mRNA and protein decay and diffusion.
We perform global parameter sensitivity analyses on these models using different initial conditions and model output at different spatial and temporal points. Through this analysis, we have begun to better understand the relative importance of each parameter, to compare these sensitivities with the experimentally measured contributions to protein abundance of the biological processes they represent, and have found that parameter sensitivities themselves are temporally dynamic.