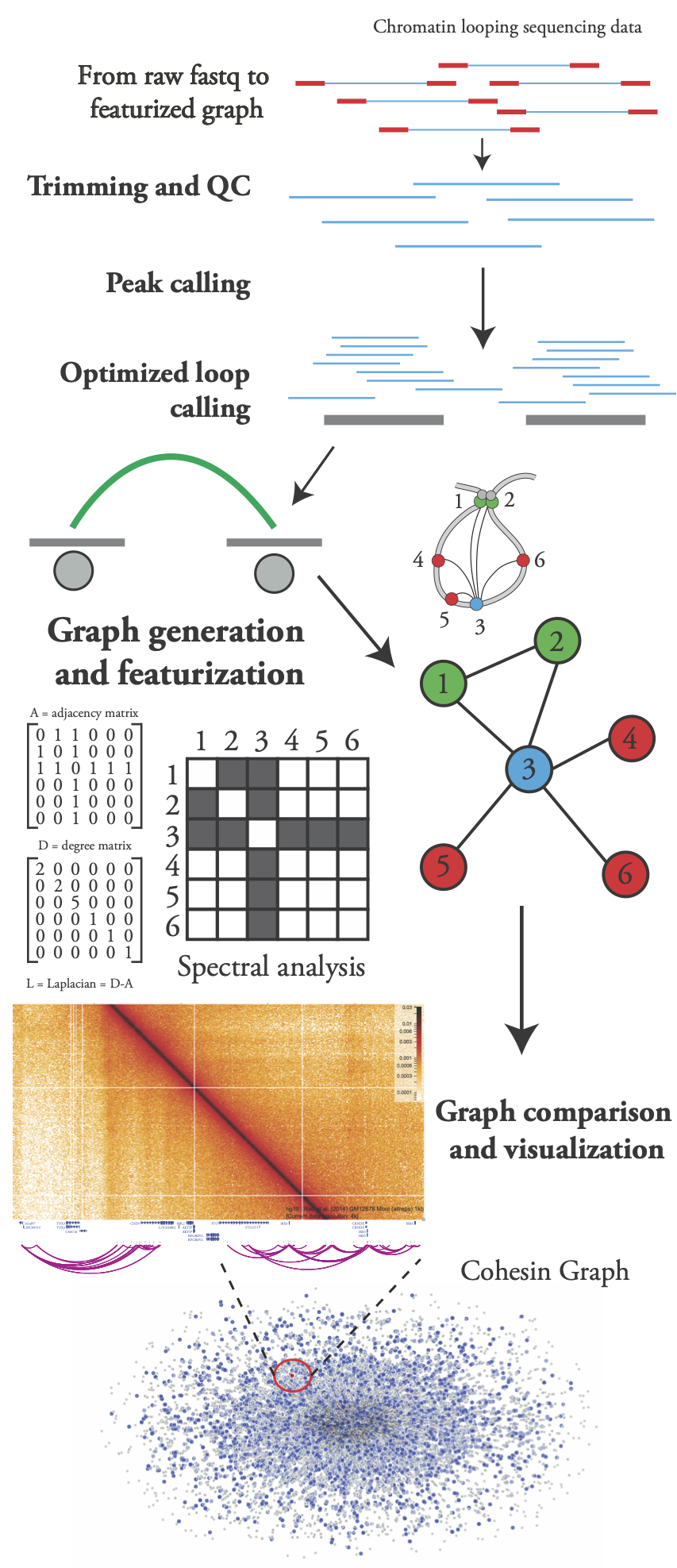
Chromatin loop extrusion shapes eukaryotic genomes, defining the physically-mediated transcriptional programs performed by the cell. A number of wet lab methods exist to measure 3D interactions in the genome, but the dry-lab mathematical framework surrounding their analysis is lacking. Our lab is working on processing chromatin looping datasets of varying resolution/quality and developing methodologies to create graphical representations of the physical shape the genome. Using methods from graph theory as well as spectral analysis, we are able to characterize and compare different representations. This approach will allow us to investigate biologically meaningful features of key developmental genes in Drosophila and dissect different modes of epigenetic regulation across evolution and development.