Our mission is to unlock the secrets of microbial population dynamics to increase plant health.
Disentangling the complexities that shape microbe-microbe-plant interactions will provide a fundamental understanding of the ecological and evolutionary mechanisms that organize microbial communities and serves as a novel method to quantify the influence of microbes on plant fitness.
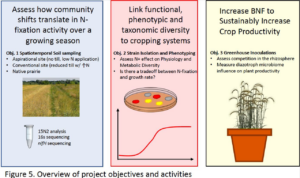
Linking functional and taxonomic diversity of diazotrophs under contrasting management regimes for enhanced crop productivity
Funded by USDA – NIFA Foundational Agriculture Award # 2021-67013-33677
This project will generate foundational knowledge using concepts in microbial evolutionary ecology to link the functional and taxonomic diversity of beneficial soil microbes to cropping systems. The project will measure how free-living nitrogen fixing bacteria (diazotroph) population dynamics respond to soil properties and agriculture management regimes, and document how community shifts translate into N-fixation activity over the growing season. We will determine whether potentially beneficial diazotrophs may have been lost through conventional agronomic management by using a prairie remnant as a baseline for community diversity. The knowledge gained could increase biological N-fixation rates as a means to decrease synthetic fertilizer dependence, which aligns with the program area priority to sustainably increase crop productivity. To accomplish our goals, we will spatiotemporally sample soils across Cook Agronomy Farm (CAF), which operates no-till and reduced till fields, to measure N-fixation and microbial communities. We will isolate free-living nitrogen-fixing strains from CAF, and characterize their metabolic diversity to assess the effect of nitrogen fertilizers. Strains will then be grown in competition with one another to predict the outcome of interactions in soil. We will test whether strains that fix more nitrogen show fitness costs and are less competitive when grown in the presence of mineral nitrogen sources, which we predict would both decrease diversity and lower fixation rates in the field. Finally, we will inoculate wheat in the greenhouse to assess competition in the rhizosphere and to measure the diazotroph microbiome’s influence on plant productivity.
Transgenerational rhizobial selection in Trifolium barbigerum plants
Funded by USDA – NIFA Foundational Agriculture Award # 2021-67013-33677
This project aims to determine if Trifolium barbigerum (clover) hosts are more likely to partner with beneficial rhizobia they have had previous exposure to, even in the presence of higher N-fixing strains. Because the plant cannot assess the nitrogen-fixing ability of the rhizobia before nodule formation, it would be in the plant’s best interest to remember which strains were most helpful. Rhizobia strains were isolated from our field site in Bodega Bay, CA. Trifolium plants were inoculated with isolated strains to determine their effect on plant growth. Once beneficial and non-beneficial strains are determined, plants will be inoculated with a familiar and unfamiliar partner to see which strain is more prominent at harvest – and whether the plants can truly remember their friends!
BRC_FIGInduced versus constitutive response of plant defenses to insect herbivores across ranges
The invasion of novel environments by plants opens up opportunities for access to previously unavailable resources while also exposing the individuals to novel environmental threats. One such example of these types of threats are insect herbivores with which an introduced plant population has not co-evolved. The shifting defense hypothesis (SDH predicts that selection pressures on defensive traits will change over the course of an invasion; specifically, it predicts a shift away from more energetically expensive compounds that deter generalists and more specialized enemies to cheaper compounds that garner more broad spectrum protection. We tested these predictions by comparing feeding preferences of two herbivores on burr clover (Medicago polymorpha) genotypes from M. polymorpha’s native range around the Mediterranean and invasive range in the North and South America–neither herbivore occurs in the host plant’s native range but both are common in North and South America. We also compared the activity of peroxidase, polyphenol oxidase, and total protein production for host plant genotypes from the two ranges.