Overview
We are employing integrated interdisciplinary approaches to examine transcriptional regulation controlled by the complex interactions between multiple protein transcription factors and DNA sequences. These experimental approaches include mathematical modeling, synthetic biology, and biophysical characterization of molecular interactions.
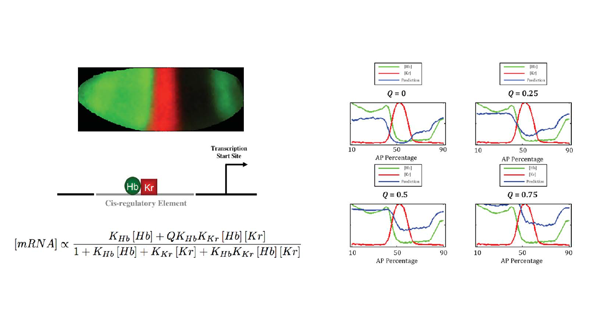
Computational modeling of TF-DNA dynamics
In the field of gene regulation, the most popular static model of transcriptional regulation for incorporating DNA level information is a thermodynamic-based (or fractional occupancy) model. Thermodynamic models can be used to predict quantitative levels of gene expression given the DNA sequence and concentration gradients of the transcription factors (TFs) involved in regulation. They do this using techniques derived from statistical physics which involve computing all possible states of TFs binding to a cis-regulatory module (CRM) and relating these states to gene expression. We are currently applying thermodynamic models of gene regulation to synthetic CRMs designed to investigate the binding strength of specific TFs and the role of certain TF interactions, such as quenching and competition, on short-range repression during early Drosophila development. The use of synthetic constructs with a small number of binding sites decreases the complexity of TF interactions, thus increasing parameter identifiability and model reproducibility.This will allow us to fit the model, with a high level of confidence, to quantitative expression data obtained from in situ hybridization of reporter genes under the control of these synthetic CRMs, and begin to uncover the nature of TF-induced regulation of gene expression.
Biophysical analysis of TF protein-DNA interactions
Homeodomain transcription factors (HD TFs) are a large class of evolutionarily conserved DNA binding proteins that contain a basic 60-amino acid region required for binding to specific DNA sites. In Drosophila melanogaster, many of these HD TFs are expressed in the early embryo and control transcription of target genes in development through their interaction with cis-regulatory modules. We have developed a streamlined expression and purification protocol to produce large yields of Drosophila HD TFs. These purified HD TFs are currently being used in biophysical experiments to structurally and biochemically characterize how and why these HD TFs bind to different DNA sequences and further probe how nucleotide differences contribute to TF-DNA specificity in the HD family.