Overview
DNA methylation, the addition of a methyl (CH3) group to a cytosine residue, is an evolutionarily conserved epigenetic mark involved in a number of different biological functions in eukaryotes, including transcriptional regulation, chromatin structural organization, cellular differentiation and development. We are investigating the biological significance of this epigenetic mark across a range of eukaryotic species.
Methylation in the Dictyostelium slime mold
In the social amoeba Dictyostelium, previous studies have shown the existence of a DNA methyltransferase (DNMA) belonging to the DNMT2 family, but the extent and function of 5-methylcytosine in the genome are unclear. In our recent studies we analyzed the whole genome DNA methylation profile of Dictyostelium discoideum using deep coverage replicate sequencing of bisulfite-converted gDNA extracted from post-starvation cells. We discovered an overall very low number of sites with any detectable level of DNA methylation. Furthermore, a knockout of the DNMA enzyme leads to no overall decrease in DNA methylation. Of the identified sites, significant methylation is only detected at 11 sites in all four of the methylomes analyzed. Targeted bisulfite PCR sequencing and computational analysis demonstrate that the methylation profile does not change during development and that these 11 cytosines are most likely false positives generated by protection from bisulfite conversion due to their location in hairpin-forming palindromic DNA sequences.
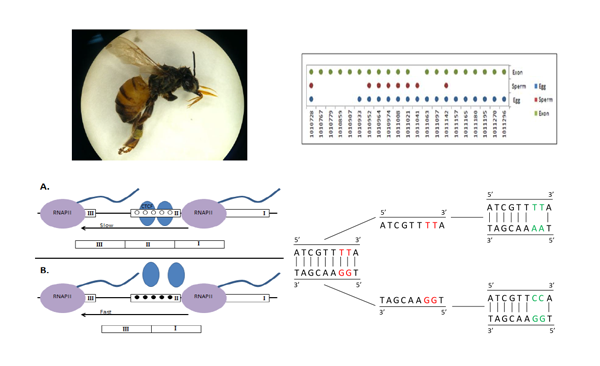
Genomic imprinting in the honey bee methylome
The Kin Theory of Genomic Imprinting (KTGI) predicts that imprinting will arise when the reproductive interests of parents differ. In colonies of the eusocial honeybee, Apis mellifera, a queen’s interests are maximized if she monopolizes reproduction in the colony and all of her workers are sterile. Males, in contrast, can increase reproductive success if some of their worker offspring are fertile.Honeybees possess a functional DNA methylation system, and methylation is known to play a role in caste determination. Parent-of-origin specific imprinting via methylation provides a candidate mechanism by which a queen may enforce sterility in her worker offspring; by epigenetically modifying genes required for fertility.We are examining the genome-wide methylation profile of unfertilized eggs and sperm in an effort to identify genes that show significantly differential methylation. Parentally-directed epigenetic modification of genes related to reproduction may be a key mechanism by which eusociality evolves and is maintained.