Overview
In multicellular organisms, development is a period in which precise gene expression is required to regulate cellular differentiation, leading to faithful production of the adult body plan. Differentiation is mediated by a complex network of genes that are controlled in large part by cis-regulatory modules (CRMs). CRMs are segments of non-coding DNA that bind transcription factors (TFs) to up- or down- regulate the expression of their target genes.
Hox genes
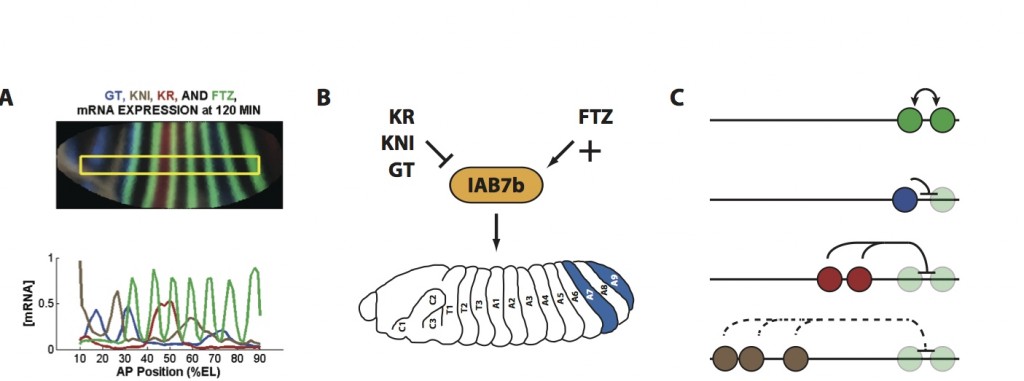
In Drosophila, cellular identity along the anterio-posterior axis falls under the control of two homeotic (Hox) gene complexes. The 330 kb bithorax complex (BX-C), which regulates cell type differentiation during development in the posterior thorax and abdomen, is comprised of three Hox genes: Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B). The expression of each of these genes is in turn controlled through interactions between transcription factors (TFs) and a number of cis-regulatory modules (CRMs) in the neighboring intergenic regions. We are investigating how the sequence architecture of TF binding sites mediates the functional activity of these CRMs using both computational and molecular genetic experimental approaches.
Cis-regulatory module function
Recent studies in Drosophila have identified minimal conserved motifs, consisting of multiple TF binding sites within CRMs, that are capable of reproducing the function of the module. These functional motifs may represent the underlying molecular mechanism by which cis-regulation drives specific gene expression patterns. We are investigating how complex combinatorial activities, such as individual protein-protein interactions, as well as the concerted effort of multiple motifs, control the functional output of a single gene. To understand the activity of these motifs and how they contribute to overall CRM function we are utilizing a synthetic biology approach, combining bioinformatic predictions, mathematical modeling and in vivo reporter gene assays in cell lines and the embryo. The goal is to functionally decode the CRM network that controls gene expression along the anterio-posterior axis in the early embryo.