Overview
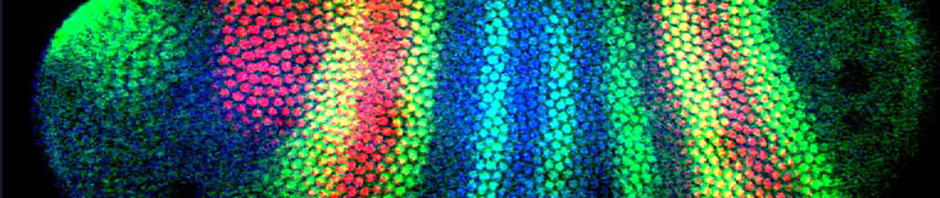
The precise regulation of gene expression is critical to early embryonic development. This transcriptional regulation is tightly controlled by the binding and complex interactions between multiple protein transcription factors (TFs) and DNA sequences within cis-regulatory modules (CRMs or enhancers). We use an integrated combination of interdisciplinary approaches, including bioinformatic, mathematical modeling, biophysical and synthetic biology experiments, and transgenic reporter assays to decipher the regulatory code of the gene regulatory networks in the fruit fly model species, Drosophila melanogaster.
Twin of eyeless
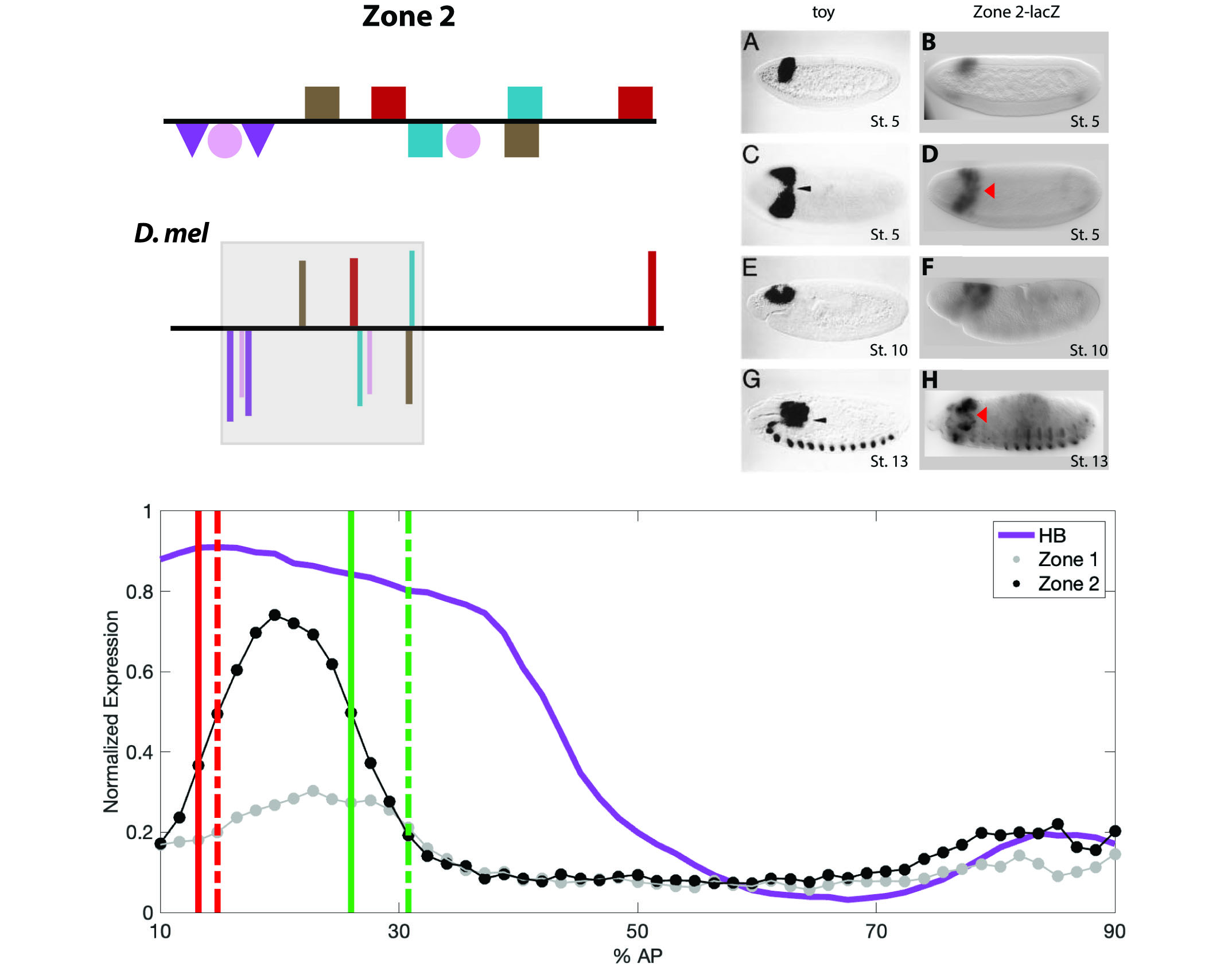
The Drosophila PAX6 homolog twin of eyeless (toy) sits at the pinnacle of the genetic pathway controlling eye development, the retinal determination network. Expression of toy in the embryo is first detectable at cellular blastoderm stage 5 in an anterior dorsal band in the presumptive procephalic neuroectoderm, which gives rise to the primordia of the visual system and brain. Although several maternal and gap transcription factors (TFs) that generate positional information in the embryo have been implicated in controlling toy, the regulation of toy expression in the early embryo is currently not well characterized. In our studies, we adopt an integrated experimental approach utilizing bioinformatics, molecular genetic testing of putative enhancers in transgenic reporter gene assays and quantitative analysis of expression patterns in the early embryo. Recently, we identified two novel coacting (shadow) enhancers at the toy gene. In addition, we apply mathematical modeling to dissect the regulatory landscape for toy and are investigating the distinct molecular architecture of the two enhancers.
Hox genes
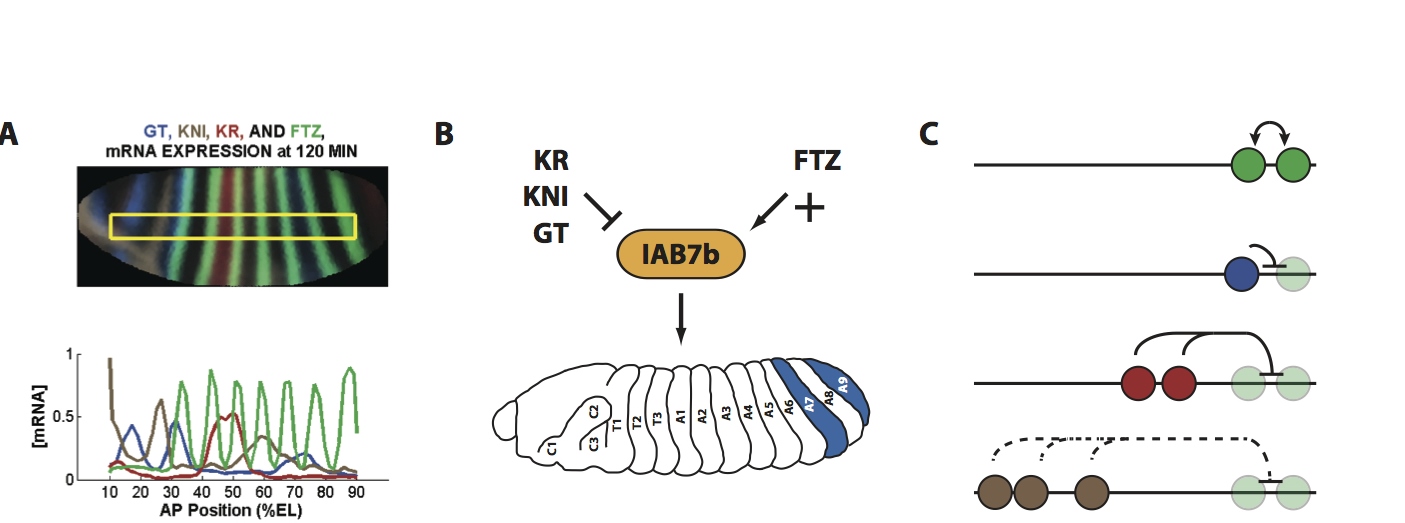
In the early Drosophila embryo, CRMs, under the control of TF concentration gradients, direct the spatio-temporal expression of a network of developmental genes, including the highly conserved homeotic genes in the bithorax complex (BX-C). The homeotic genes are critical for differentiation and specification of many important structures in development. Our ultimate goal is to elucidate the molecular mechanisms that control functional activity of homeotic gene CRMs at the sequence level, through the application of a systems-level approach. We are currently addressing the molecular contribution of individual TF binding sites within known CRMs, investigating their combinatorial interactions, and analyzing their in vivo activity. Specifically, we are deciphering the mechanisms at play in the CRMs, including the functional role of binding site affinity, the specificity and distance-dependence of interactions between repressor and activator TFs, and the role for distance-dependent cooperativity between activator proteins.